Running SIMION on HPC Clusters¶
There are some possibilities for using high performance computing (HPC) clusters to quicken your SIMION calculation workloads. SIMION presently has Multicore CPU support for running multiple threads on a node (computer) but does not natively support a formal method like MPI for spreading a computation across multiple nodes (multiple processes). Still, you can of course start up multiple independent instances of SIMION, each on its own node, if you can split up your calculation in that manner. (The License for SIMION allows this.) It is even possible of course for Lua code in SIMION instances to communicate via simple file drop, child processes/pipes or TCP sockets (see SIMION Example: multiprocess and SIMION Example: extension).
If you don’t own your own cluster, there are various low cost options to pay for on-demand remote access to a cluster at an hourly or monthly usage rate (see options below).
Ways to Effectively Use Multiple Nodes¶
The follows are some cases where it may be helpful to spread a calculation across multiple nodes in a cluster.
Refining PA# Across Multiple Nodes¶
Each solution array (e.g. .PA1, .PA2, .PA3, etc.) from a PA#
file can be solved/Refined independently, so it can be easy to
spread this calculation across multiple nodes in a cluster.
For example, if a PA# file has three adjustable electrodes, you might
refine each adjustable electrode solution array on its own node,
with a 10 or so cores per node (3*10=30 cores total).
So, rather than doing simion --nogui refine --convergence 1e-7,
you can run the following commands separately on three or four nodes:
simion --nogui lua -e "local pa = simion.pas:open('quadout.pa#'); pa:refine{solutions={0}, convergence=1e-7}"
simion --nogui lua -e "local pa = simion.pas:open('quadout.pa#'); pa:refine{solutions={1}, convergence=1e-7}"
simion --nogui lua -e "local pa = simion.pas:open('quadout.pa#'); pa:refine{solutions={2}, convergence=1e-7}"
simion --nogui lua -e "local pa = simion.pas:open('quadout.pa#'); pa:refine{solutions={3}, convergence=1e-7}"
Note: In contrast to the other files, the .PA0 file (solution=0) is created quickly if all electrodes are fast adjustable (no .PA_ file), so you may or may not want to create it on yet an another node.
Flying Particles on Multiple Nodes¶
Simulations having a large number of particles (e.g. Monte Carlo Method) can be benefit by splitting the particle trajectories across multiple instances of SIMION, assuming the particles are independent (e.g. not using ion-ion charge repulsions). For example:
simion --nogui fly --particles=1.ion example.iob
simion --nogui fly --particles=2.ion example.iob
simion --nogui fly --particles=3.ion example.iob
Optimization¶
You could structure an Optimization problem
(e.g. SIMION Example: geometry_optimization or SIMION Example: tune),
to run each parameterization as a separate fly’m
on a different core or node.
Just schedule a simion --nogui fly example.iob command
for each parameterization you wish to run.
You may differentiate the runs by passing adjustable variables
or Lua global variables:
simion --nogui fly adjustable n=1 example.iob
simion --nogui fly adjustable n=2 example.iob
simion --nogui --lua "n=1" fly example.iob
simion --nogui --lua "n=2" fly example.iob
Linux Clusters¶
Many clusters are Linux only, so you will need to either run the non-GUI native Linux SIMION binaries or the GUI SIMION under Wine32 or Wine64 (if Wine is supported or can be compiled on the cluster). You might even use a combination of this: the Linux binaries for computation workloads and the Windows binary GUI binary for pre- and postprocessing.
Data Transfer¶
Some SIMION files like potential arrays (PA) can be quite large (e.g. on
the order of a gigabyte).
If you are doing your pre or post processing on a computer separate
from the cluster (e.g. a Windows desktop), you may need to transfer
large files, so a fast network connection is preferable,
although this is more challenging if using a relatively
slow Internet connection.
One way to address this is to do more of your pre- or post-processing
remotely on the cluster itself.
Another approach is to reduce the amount of data transfer.
Note that unrefined potential arrays compress well by zipping them.
You can also crop refined potential arrays if only certain regions
are still needed after refining
(Modify Crop or (simion.pas pa:crop().
If data transfer remains an issue, contact us about possible
other techniques.
Service Providers Offering On-Demand Access to HPC Nodes¶
One company offering on-demand HPC services, which SIMION has some familiarity with and which we would be glad to work with you with and perhaps more officially support in the future, is Sabalcore http://www.sabalcore.com/. As of 2014, the cost was roughly on the order of up to USD$0.25 per core hour (e.g. six hours on 16 cores was USD$24), with little or no additional cost for bandwidth or disk access. The clusters have many nodes, many cores per node, and lots of RAM per core, with fast communication and shared disks between the nodes. Systems are Linux based. Job scheduling on the clusters is done via PBS TORQUE. Systems can be accessed via a remote login to a graphical desktop launched from a web browser (Java app with 3D acceleration) or SSH. See the Sabalcore documentation for the latest specifications.
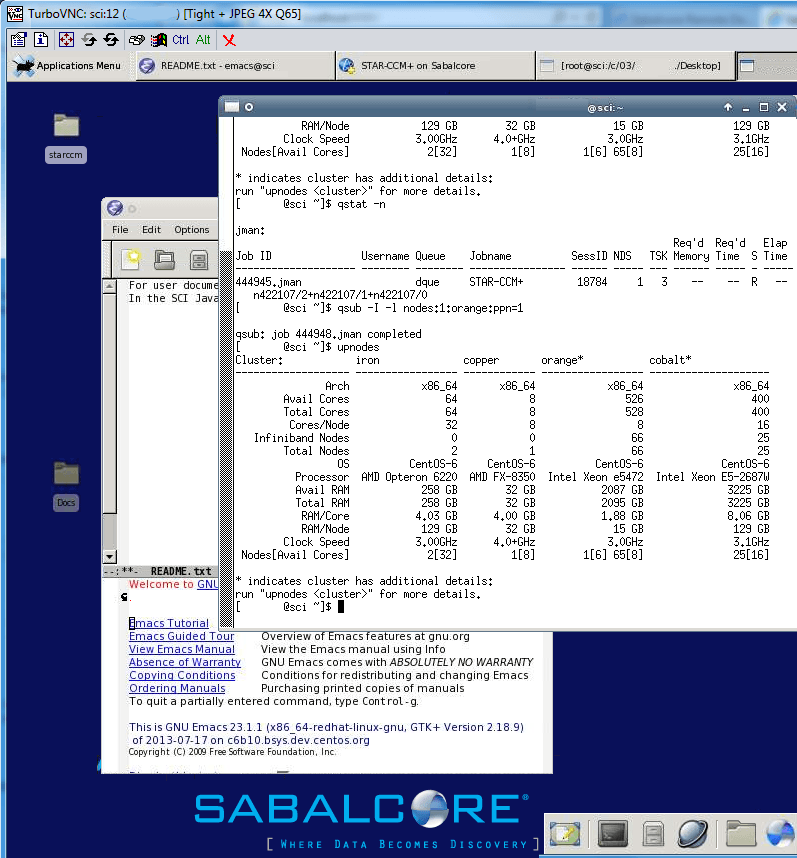
Fig. 71 Logged into graphical desktop of Sabalcore cluster
with web-based Java client and viewing cluster notes (upnodes)
and job queue status (qstat).¶
Another option is SIMION on Amazon EC2. Sabalcore has said that their offering compared to EC2 is more of a traditional, high-end HPC cluster for numerical work, with the expected fully managed/preinstalled HPC tools and schedulers, fast (low latency) inter-node communication (Infiniband), and physical (not virtualized) hardware. They are also differentiated on comprehensive technical support and have experience running and managing other scientific simulation codes like for Computational Fluid Dynamics (CFD). EC2 does offer Windows.
Other links
Other Notes¶
This documentation is preliminary and should be expanded in the future. If you are running clusters with SIMION, we could be glad to hear your feedback.